

Stilla Technologies Digital PCR System
naica®Crystal Digital PCR System
NaicaTMCrystal Digital PCR System
Azure Biosystems Real-Time PCR Systems

Considering the highly dynamic nature of mRNA transcription and the potential variables introduced in the sample processing and downstream processing steps, a standardized method for each step of the RT-qPCR workflow is essential for reliable and reproducible results. MIQE provides a list of 85 parameters for this method to ensure that the quality results meet the acceptance criteria of any journal. Next, we will introduce in detail how to use the MIQE guide to establish a reliable RT qPCR experimental process.
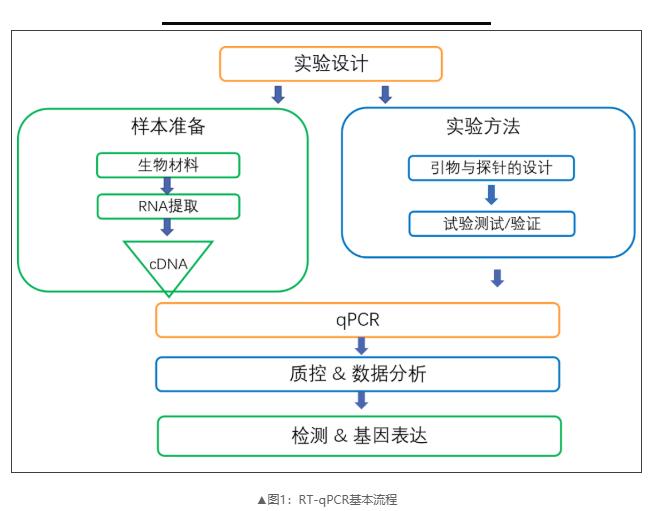
1、Experimental Design
Correct experimental design is the key to any gene expression research. Because mRNA transcription is sensitive to external stimuli unrelated to the research process, it is very important to strictly control and design the experimental conditions. This includes the determination of experimental procedures, the type and number of control and repeat groups, experimental conditions, and sample handling methods within each group, which are essential to minimize variability (Table 1).

2、RNA Extraction and Quality Control
Samples should be frozen at -80 ℃ or stored in RNA storage solution. RNA extraction procedures should include DNA enzyme treatment steps to remove any genomic DNA contamination.
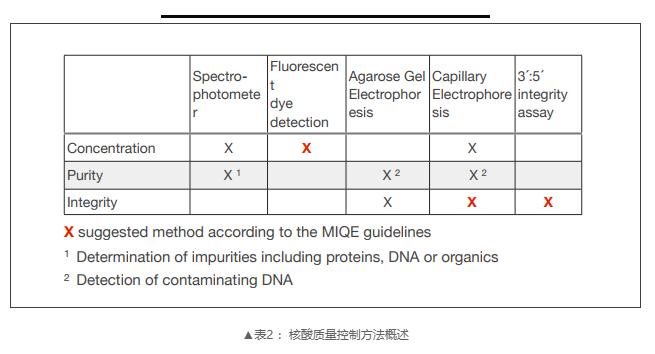
To ensure that only high purity (contaminant free) and high integrity (undegraded) RNA are used is the most critical point in the RT qPCR experimental workflow. Impurities in RNA samples may lead to inhibition of RT and PCR, resulting in different and incorrect quantitative results. Since sample purity and integrity are not related, both should be evaluated to determine that RNA samples meet the minimum acceptance criteria for downstream workflow.
3、Reverse Transcription
Considering the widespread presence of RNase in the environment, it is recommended that the total RNA sample be reverse transcribed into cDNA immediately after the quality control evaluation. This will avoid the risk of RNA sample degradation due to multiple freezing / thawing before transforming into cDNA. For RT, the key is to ensure the consistency and integrity of the transcriptional genome in the extracted RNA samples. It is recommended to use the same amount of total RNA and keep the reverse transcription reaction time of all experimental samples to minimize the variability between biological replications.
4、Design of Primers and Amplicons
Primer design and target sequence selection are the key to ensure the specificity and high efficiency of amplification products. At present, there are many softwares or online websites that can design primer pairs and determine amplicons, such as DNAMAN、Primer Premier、Oligo、Beacon Designer、Primer-Blast、BathPrimer、Primer3 Plus, etc.
5、qPCR Validation
qPCR validation is a method to evaluate the annealing temperature, reaction efficiency and the appropriate range of specificity of primers using a set of standard samples. The specific steps are as follows:
1) Appropriate consumables and qPCR reagents were selected according to the type of instrument;
2)different PCR reaction systems were configured, and the suitable concentration and specificity of primers were detected by gel electrophoresis;
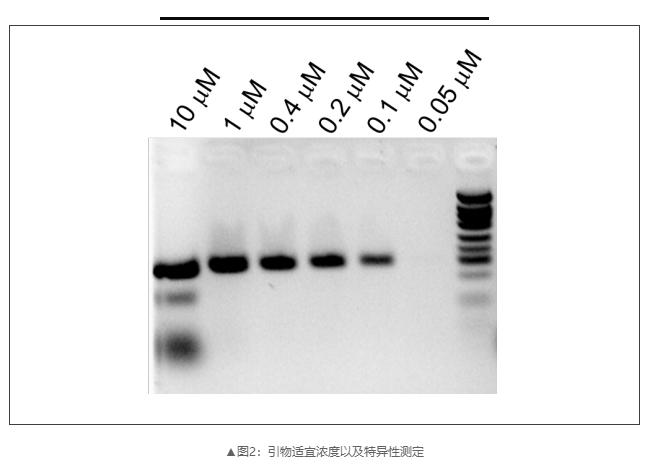
3)The suitable annealing temperature was set for the temperature gradient test primer;

4)NTC, NRC, POS, neg and other control groups were set to monitor the experimental system or pollution;
5)Establishment of standard curve (evaluation of PCR efficiency)。

6、Selection of Internal Reference Genes
In the RT-qPCR experiment, the internal reference gene can be used to correct the experimental errors in the loading amount and the loading process, and ensure the accuracy of the experimental results. A good internal reference gene should have a relatively stable expression level in different time and space samples or different treatment samples, such as ATCB, GAPDH, β - actin, 18S rRNA and so on.
7、Repeatability of Experiments
There are two possible sources of variation in gene expression experiments:
1)Biological variability due to inherent differences in gene expression levels between individual organisms, tissues or cell samples;
2) The technical variability of the experimental process itself is usually related to pipette error, operation error or sample quality and quantity. In order to reduce the impact of biological and technical variability, it is generally believed that at least three biological and technical repetitions are needed.

We have seen the whole RT-qPCR experimental process. Have you lifted the veil of mystery of RT qPCR? Now come to apply for Azure Cielo™ Real-Time PCR System to feel the real and reliable RT qPCR data it brings to you.
If you want to know more about it, please download the new version of MIQE guide 2020: http://www.econferences.de/miqe-qpcr-ibook/
Azure Cielo™ Real-Time PCR System
Azure Cielo™ Real-Time PCR System is from Azure Biosystems company of the United States, which can provide 3 / 6 detection channels for you, and can be flexibly configured according to the experimental requirements. This product uses high-energy LED as the light source system, which can ensure high light intensity and good light source consistency; high quality Peltier temperature module as the temperature control system, with fast temperature rise and fall speed, which can set 12 columns of 30 °C span temperature gradient; excellent CMOS photography + optical fiber signal transmission as the detection system, with high CMOS detection sensitivity, fast optical fiber transmission speed, no light loss and noise, there is no need for Rox calibration. Azure Cielo™ Real-Time PCR System can provide experimental results with high accuracy, high sensitivity and high reliability for your scientific research.

