

Stilla Technologies Digital PCR System
naica®Crystal Digital PCR System
NaicaTMCrystal Digital PCR System
Azure Biosystems Real-Time PCR Systems

Real-time PCR technology is based on conventional PCR by adding fluorescent labeled probes or corresponding fluorescent dyes to achieve its quantitative function. Because of its good accuracy and high sensitivity, it has been widely used in molecular biology and medical research, especially in gene expression difference analysis. Gene expression difference analysis is generally used to compare the expression differences of specific genes between different spatiotemporal samples or different treatment samples (drug treatment, physical treatment and chemical treatment, etc.).

When we detect that the expression of a gene is up-regulated or down regulated, we do not know whether the expression difference is due to the change of its own expression, or the sample size or RNA loss caused by operation and other reasons. In order to reduce experimental bias and produce accurate expression level, RT-qPCR often needs to consider several variables (such as operation error or RNA extraction amount, yield and variation) for normalization. At present, the most common method of RT-qPCR standardization is to use internal reference genes for homogenization.
What are internal reference genes?
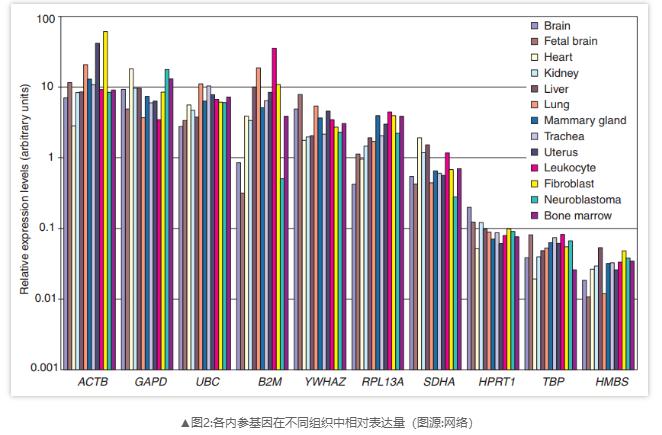
Ideally, the expression level of internal reference genes should be relatively stable in all samples of different tissues at all developmental stages and in response to different experimental treatments. Internal reference genes are usually various housekeeping genes, such as ATCB, GAPDH, β - actin, 18S rRNA, etc. Their expression level is less affected by environmental factors, and can be continuously expressed in almost all tissues and all growth stages of organisms. In different tissues, housekeeping gene mRNA are abundant.
Function of internal reference gene
In gene expression difference analysis, internal reference genes can be used to correct the experimental errors in sample loading and sample loading process, so as to ensure the accuracy of experimental results. With the help of testing the amount of internal reference of each sample, it can be used to correct the loading error, so that the quantitative results are more reliable. Generally, a gene that will not change expression under the condition of treatment factors should be selected as internal reference.
The ideal internal reference gene should meet the following conditions:
1.There is no pseudogene to avoid the amplification of gene DNA;
2.High or moderate expression, excluding low expression;
3.It was stably expressed in different types of cells and tissues (such as normal cells and cancer cells), and its expression level was similar without significant difference;
4.The expression level was not related to cell cycle and activation;
5.Its stable expression level is similar to the target gene;
6.Not affected by any endogenous or exogenous factors, such as not affected by any experimental treatment measures.
Evaluation of internal reference genes
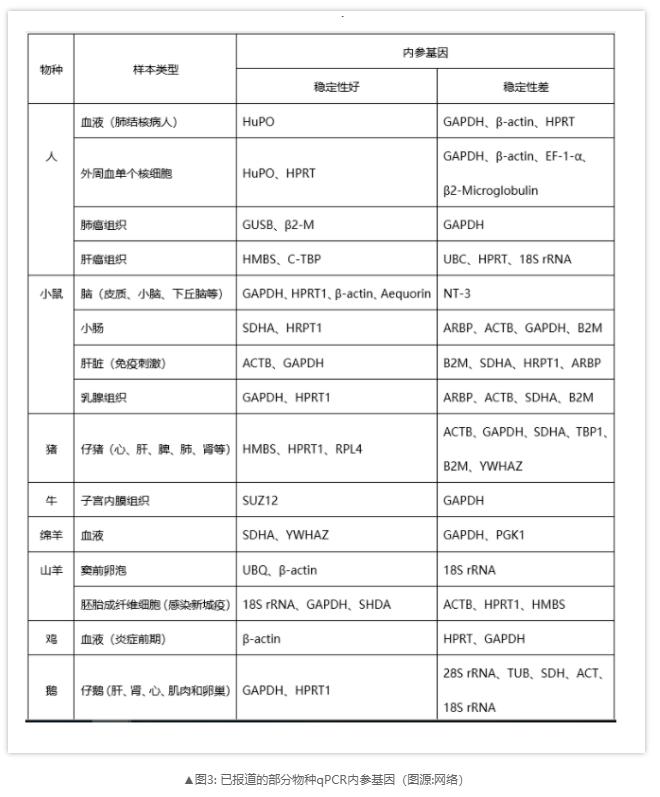
However, there is evidence that some of the traditional reference genes used to control experimental bias are relatively constant expression levels only under specific conditions. Obviously, the internal reference gene has a certain specificity, and there is no universal internal reference gene that can be used for internal control of all experiments, which also indicates that the internal reference gene must be selected correctly before RT qPCR experiment. When selecting internal reference genes, we can use tools such as geNorm, BestKeeper, NormFinder and refgenes to evaluate, which is the premise to ensure the reliability and accuracy of the results. Secondly, the candidate internal reference genes can be further screened by qPCR experiment. The internal reference gene stably expressed in different samples is preferred, that is, the average value of Cq and the standard deviation of Cq value in each sample are compared, and the gene with the smallest SD is selected as the internal reference.

Do you know about the selection of internal reference genes now?
Azure Cielo™ Real-Time PCR System
Azure Cielo™ Real-Time PCR System is from Azure Biosystems company of the United States, which can provide 3 / 6 detection channels for you, and can be flexibly configured according to the experimental requirements. This product uses high-energy LED as the light source system, which can ensure high light intensity and good light source consistency; high quality Peltier temperature module as the temperature control system, with fast temperature rise and fall speed, which can set 12 columns of 30 °C span temperature gradient; excellent CMOS photography + optical fiber signal transmission as the detection system, with high CMOS detection sensitivity, fast optical fiber transmission speed, no light loss and noise, there is no need for Rox calibration. Azure Cielo™ Real-Time PCR System can provide experimental results with high accuracy, high sensitivity and high reliability for your scientific research.

