

Stilla Technologies Digital PCR System
naica®Crystal Digital PCR System
NaicaTMCrystal Digital PCR System
Azure Biosystems Real-Time PCR Systems

NSCLC
Lung cancer is one of the most common malignant tumors in the world, and has become the first cause of death from malignant tumors in China's urban population. Among them, non-small cell lung cancer accounts for about 80% of all lung cancers, and about 75% of patients are already in the middle and late stages when found, with a low 5-year survival rate.
In patients with non-small-cell lung cancer (NSCLC), the possibility of treatment with tyrosine kinase inhibitor (TKI) is determined by the presence of mutations on exons 18–21 of the epidermal growth factor receptor (EGFR).1 Clinical evidence heightens the fact that EGFR-targeted therapy can significantly improve progression-free survival (PFS) and overall survival (OS) in patients.2,3 Therefore, assessing the status of EGFR mutations is important for potential . The most common mutation in non-small cell lung cancer that has targeted drugs is the "epidermal growth factor receptor" (EGFR) mutation. The EGFR gene is located on the short arm of chromosome 7 and is a tyrosine kinase receptor. Mutations in the EGFR tyrosine kinase region occur mainly in exons 18 to 21 (see figure below). Therefore, the early detection of EGFR mutations in NSCLC patients will allow for targeted treatment to reduce patient suffering and prolong the life cycle.
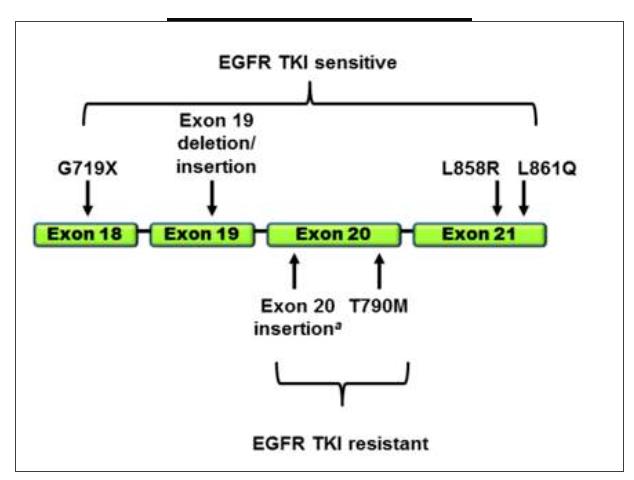
▲ EGFR-TKI therapy
Digital PCR, as a third-generation PCR technology, is becoming a reliable method for noninvasive plasma testing-liquid biopsy by virtue of its high sensitivity, high precision, and strong tolerance to inhibitors. However, factors such as low throughput and high cost of digital PCR in most cases have limited its application in clinical testing.
The research and development team at Apexbio recently published an article entitled 《Plasma-based early screening and monitoring of EGFR mutations in NSCLC patients by a 3-color digital PCR assay in the prestigious journal British Journal of Cancer》 (BJC). It provides a highly sensitive digital PCR technique to detect mutations in plasma levels of EGFR in NSCLC patients, and can simultaneously detect 39 mutations in exons 18-21 of the EGFR gene simultaneously, greatly improving clinical EGFR mutation detection, especially when the samples to be tested are limited, and this method can obtain more genetic information from a small number of samples. The impact factor of the journal is 5.791. (Please see the end of the article for more details)
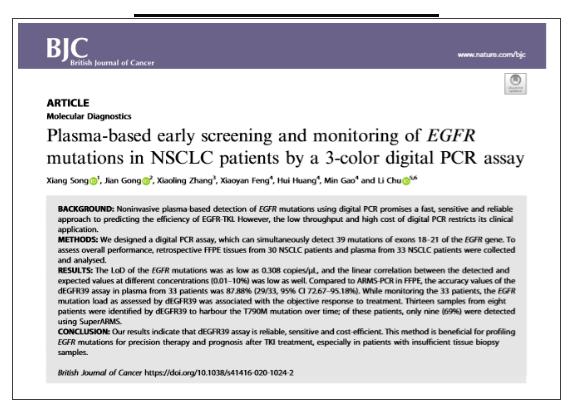
BACKGROUND:
Noninvasive plasma-based detection of EGFR mutations using digital PCR is expected to be a rapid, sensitive and reliable approach to predicting the efficiency of EGFR-TKI. However, the low throughput and high cost of digital PCR limit its application in clinical application.
METHODS:
We designed a digital PCR method, which can simultaneously detect 39 mutations of exons 18–21 of the EGFR gene. For a more comprehensive evaluation, retrospective FFPE tissues from 30 NSCLC patients and plasma from 33 NSCLC patients were collected for analysis.
RESULTS:
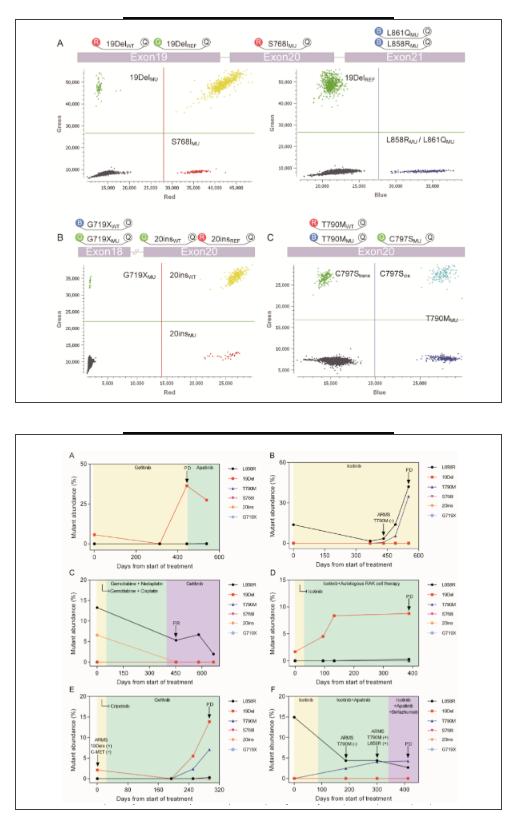
The LOD of the EGFR mutations was as low as 0.308 copies/μL, and the linear correlation between the detected and expected values at different concentrations (0.01–10%) was low as well. Compared to ARMS-PCR in FFPE, the accuracy values of the dEGFR39 assay in plasma from 33 patients was 87.88% (29/33, 95% CI 72.67–95.18%). While monitoring the 33 patients, the EGFR mutation load as assessed by dEGFR39 was associated with the objective response to treatment. Thirteen samples from eight patients were identified by dEGFR39 to harbour the T790M mutation over time; of these patients, only nine (69%) were detected using SuperARMS.
In NSCLC patients with resistance to TKI treatment, an important mechanism of primary resistance involves a mutation in T790M that blocks the binding of TKI with the adenosine triphosphate domain of EGFR of the 33 enrolled patients with NSCLC, 13 samples from eight patients were identified by dEGFR39 to harbour the T790M mutation over time; of these, only nine (69%) were detected using SuperARMS. In the plasma of patient P-15, L858R and T790M mutations were simultaneously detected after 368 days of Icotinib treatment. SuperARMS PCR, however, failed to detect the T790M mutation; moreover, there was no significant progress evident in imaging until 121 days. Similarly, in patient P-25, dEGFR39 results showed an increase in mutant abundance in 19Del and T790M after 251 days of TKI treatment, which was 43 days ahead of that of imaging progression. In patient P-23, the emergence of L858R was initially observed and dropped upon Icotinib treatment. Meanwhile, T790M was detected by dEGFR39, which gradually increased with Icotinib as well as Apatinib plus chemotherapy treatment. In this case, detection of T790M was comparatively delayed by superARMS PCR and CT imaging. Taken together, this data supports the conclusion that detection of T790M mutations by dEGFR39 occurs relatively earlier than by superARMS PCR and CT imaging, which can play a role in diagnosis and prognosis.
CONCLUSION:
Our results indicate that dEGFR39 assay is reliable, sensitive and cost-efficient. This method is beneficial for profiling EGFR mutations for precision therapy and prognosis after TKI treatment, especially in patients with insufficient tissue biopsy samples.
Naica™ Crystal Digital PCR
Crystal Digital PCR™ is Stilla®’s next-generation technology for absolute quantification of nucleic acids. Using cutting-edge microfluidic innovations, this technology integrates the Digital PCR process in a single consumable.Crystal Digital PCR™ is Stilla®’s next-generation technology for absolute quantification of nucleic acids. Using cutting-edge microfluidic innovations, this technology integrates the Digital PCR process in a single consumable.

